Reverse and find complement sequence in R
 Photo by
Emile Perron on
Unsplash
Photo by
Emile Perron on
UnsplashRecently, I am continuously being amazed by how a seemingly simple task is actually implemented in a sophisticated way. I guess I am just taking so many things for granted just because it was implemented and refined to an extent that I don’t even feel it.
When someone asked me “how do you reverse and then find the complement sequence of some DNA?” I googled and found a couple of functions, and then I decided to challenge myself to re-invent this wheel.
My goal in this post is to build is a function that:
- Reverse the sequence
- Convert base by base: ATCG to TAGC for DNA; AUGC to UACG for RNA
Since R treats the character type as a whole and does not provide an
explicit way to reverse it, I start from building one function that:
- Split a multi-charactered string into a vector of alphabets
- Reverse it with
rev()
seq_rev <- function(char) {
alphabets <- strsplit(char, split = "")[[1]]
return(rev(alphabets))
}
- Find complementary nucleotides.
seq_compl <- function(seq) {
# Check if there's "T" in the sequence
RNA <- Reduce(`|`, seq == "U")
cmplvec <- sapply(seq, function(base) {
# This makes DNA the default
# As long as there's no U, the sequence is treated as DNA
if (RNA) {
switch(base, "A" = "U", "C" = "G", "G" = "C", "U" = "A")
} else {
switch(base, "A" = "T", "C" = "G", "G" = "C", "T" = "A")
}
})
return(paste(cmplvec, collapse = ""))
}
Since I want to accommodate both DNA and RNA, one extra thing I did here was to distinguish them by the presence of uridine (“U”) in the sequence, which is specific to RNA.
Now, I have functions for reversal and complementation. Let’s test if it works .
seq_compl(seq_rev("CCAAT"))
> seq_compl(seq_rev("CCAAT"))
[1] "ATTGG"
It looks okay, but it’s in fact failure prone. You might notice that my script assumed the sequences to be in upper case, which might not always be true. Additionally, if anything other than the bases, like a number or “X”, is passed to the functions, the functions do not know how to handle them.
So, I’ll write a function that:
- Wrap the two naive functions from above.
- Handle the issues that are foreseeable.
revcom <- function(input) {
# Make sure the input is character and in upper case
input <- as.character(input)
input <- toupper(input)
# Use regular expression to check if there's only legal bases
# present in the sequence
legal_char <- Reduce(`&`, grepl("^[A,T,C,G,U]*$", input))
if (!legal_char) {
stop("revcom() only applies to DNA/RNA sequences, and only A/T/C/G/U is allowed")
}
rev <- seq_rev(input)
return(seq_compl(rev))
}
The revcom() works like this now:
# The function seems to work
> revcom("ATTTCAT")
[1] "ATGAAAT"
# It handles illegal characters and provides an error
> revcom("ATTTCATX")
Error in revcom("ATTTCATX") :
revcom() only applies to DNA/RNA sequences, and only A/T/C/G/U is allowed
# Non-character input is converted to character and then judged
# whether they are legit
> revcom(1)
Error in revcom(1) :
revcom() only applies to DNA/RNA sequences, and only A/T/C/G/U is allowed
Ok, I’ve made my “wheel”. It’s time to see if the wheel rolls well. Considering how long genomic sequences can be, I chose running speed as the benchmark here.
# Generate a 100 kb random DNA sequence
testseq <- paste(sample(x = c("A", "T", "C", "G"), replace = TRUE, size = 1e5),
collapse = "")
# Preallocate a vector to save the running time for each repeat
elapsedtime <- vector()
# Find reverse-complement sequence for 10 times
for (i in seq(10)) {
start <- Sys.time()
invisible(revcom(testseq))
end <- Sys.time()
elapsedtime[i] <- end - start
}
# Print the averge time spent
print(mean(elapsedtime))
[1] 0.1781383
Though the way I look for legal bases and that grepl() I used to check
if a sequence is RNA posed extra computation on the function, a hundred
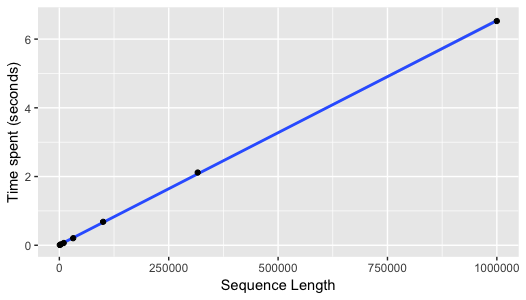
thousand bases in 0.18 seconds feels reasonably fast. Converting several
sequences with their length ranging between 1 kilo bases to 1 mega bases
shows the time required for conversion increases linearly with sequence
length. It seems to make sense, since I every base is examined and
converted independently, so no extra work should be introduced with a
longer length.