Downloading multiple samples from a entry at GEO Dataset
Openly shared data is invaluable. It provides a way for others to test reproducibility of analysis and reduces the need of repeated screening experiments. Besides, these data is also an excellent training ground for amateurs like me.
Sometimes, the dataset I want consists of multiple samples. I first clicked all the download links manually, but I soon got lost and forgot which ones I hadn’t downloaded. Thankfully, I realized repetitive tasks like this on a computer can often be automated.
For a dataset with multiple experiments, like this one, I often want to get all the raw data or the supplementary summary tables, and I am going to describe how I did it here.
Batch download of supplementary tables
In the bottom of the GSE series page, there are several files containing
metadata of the series. Each of these files contain the details about
the experiments, and every entry is in the form of ![feature name] = feature description. Luckily, these metadata all provide a list of
downloading list.
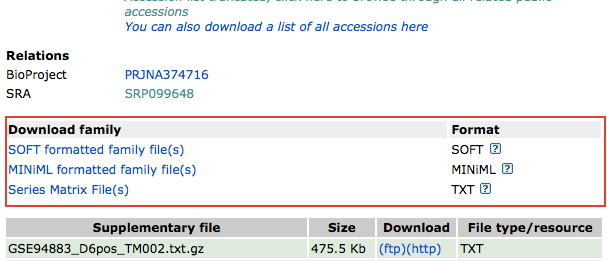
Link for each supplementary file is provided
as !Series_supplementary_file in these files, so we can use regular
expression in awk to find the link. The line looks like
this: !Series_supplementary_file = ftp://ftp.ncbi.nlm.nih.gov/geo/series/GSE94n nn/GSE94883/suppl/GSE94883_D6pos_TM002.txt.gz.
In awk, regular expression operator is ~, so $0 ~ "^!Series_supplementary_file” finds the lines containing links. All
these lines start with !Series_supplementary_file = , which we don’t
want and is consisted of 29 characters. Here, substr() can help us
removing these 29 and leaves only the link.
# This command shows all the links in terminal
awk '$0 ~ "^!Series_supplementary_file" {print substr($0, 30)}' GSE94883_family.soft
If your machine has gawk installed, the command above should print the
links for you, and the last step we need is to pass those to a download
command. I use curl for downloading in command line, so I need find a
way for awk to communicate with system, and send the links to curl.
The function that helps us using system command in awk is system(),
and with its power, we are able to pass the link to curl -O and let
the computer do the rest of downloading.
awk '$0 ~ "^!Series_supplementary_file" {system ("curl -O " substr($0, 30))}' GSE94883_family.soft
Batch download of raw data
What if I want raw data, so I could do analysis from scratch? The
metadata also gives you a SRA accession so you can access those. The
link to start with is saved as !Series_relation. The link will lead
you to a page that list all the SRA results, and this page contains a
link to Run selector, where you can download an accession list. Every
accession number is listed in this file.
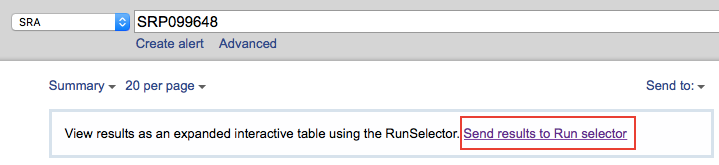
With the list (and
SRA-tools), you
should be able to download raw data as .fastq (or other file format
SRA-tools support) with a little help with awk.
awk '{system ("fastq-dump " $0)}' SRR_Acc_List.txt
Then, we are ready for analysis.