Resource usage to deduplicate BAM files with UMI-tools
While having access to HPC, resource is not infinite, and our HPC reasonably manage job queues by how much computation you have requested recently. This provides incentive to only request enough for whatever task you work on, but for me, it hasn’t been easy: Estimating resource usage is not trivial, and trying to be as frugal as possible would sometimes result in time-consuming jobs that are terminated after several hours or days due to insufficient memory or running time requested.
Therefore, try to keep an record of things that I do repetitively seems like a good idea.
umi-tools
I have been mainly using it to de-duplicate reads from BAM files generated by
cellranger, so the command that I use is usually like:
# De-duplication with umi_tools.
# Errors could happen due to resource limit and might need to adjust manually.
umi_tools dedup --extract-umi-method=tag \
--umi-tag=UB \
--cell-tag=CB \
--stdin=${path_to_bam_to_dedup} \
--log=${path_to_save_dedup_log} > ${path_to_save_deduped_bam}
As you can see from my comment, I encountered errors more than often so I had to remind myself. Recently, I finally took this seriously enough to examine how resource usage scales.
umi_tools dedup scales linearly with input file size
In short, both max RAM usage and running time scaled linearly with the size of the BAM file to de-duplicate.
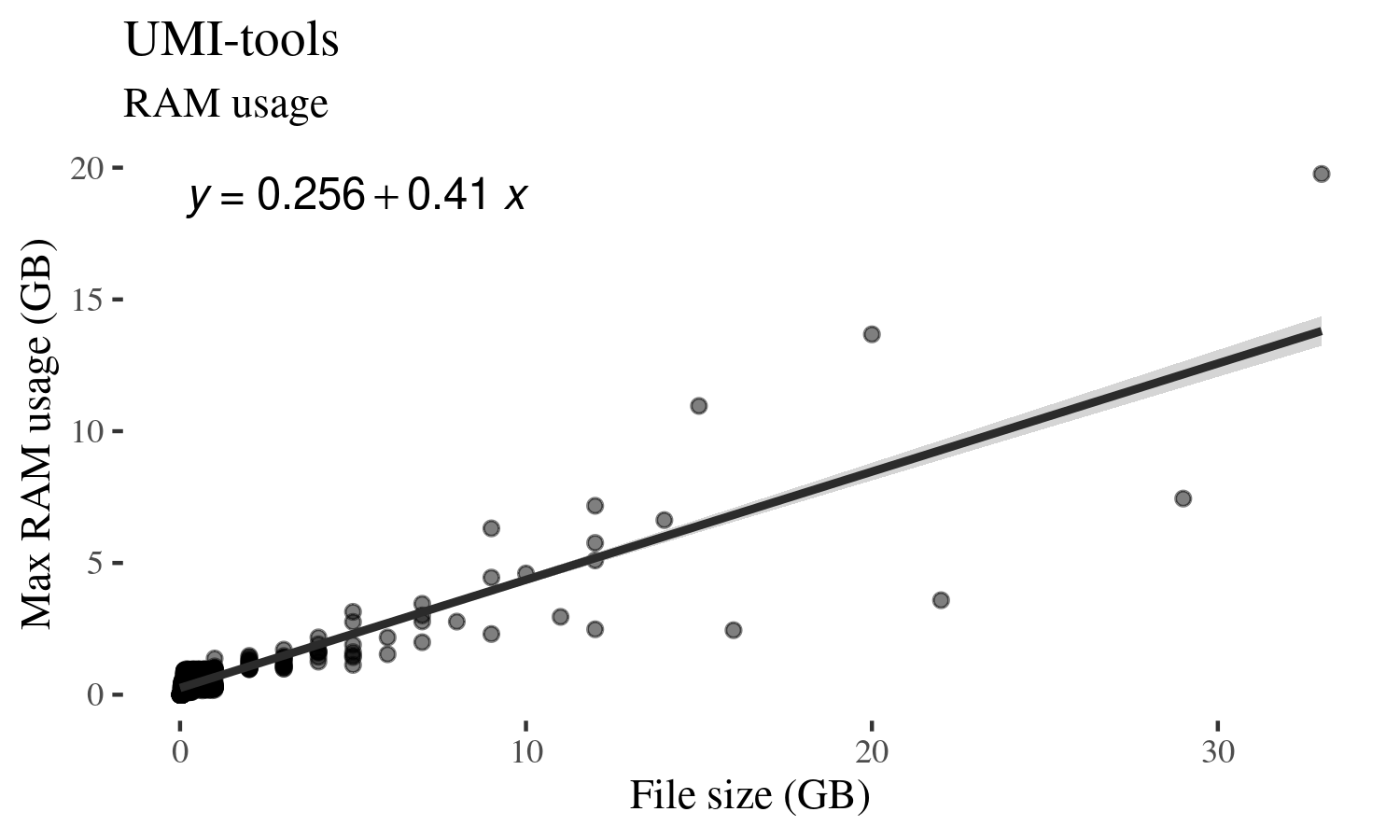
You can see above that while there are variations, input file size and max RAM
usage during run shows a linear relationship. The variation could reflect how
many UMIs were mapped to the same genomic location as that would significantly
change the complexity of UMI graph that umi-tools generates.
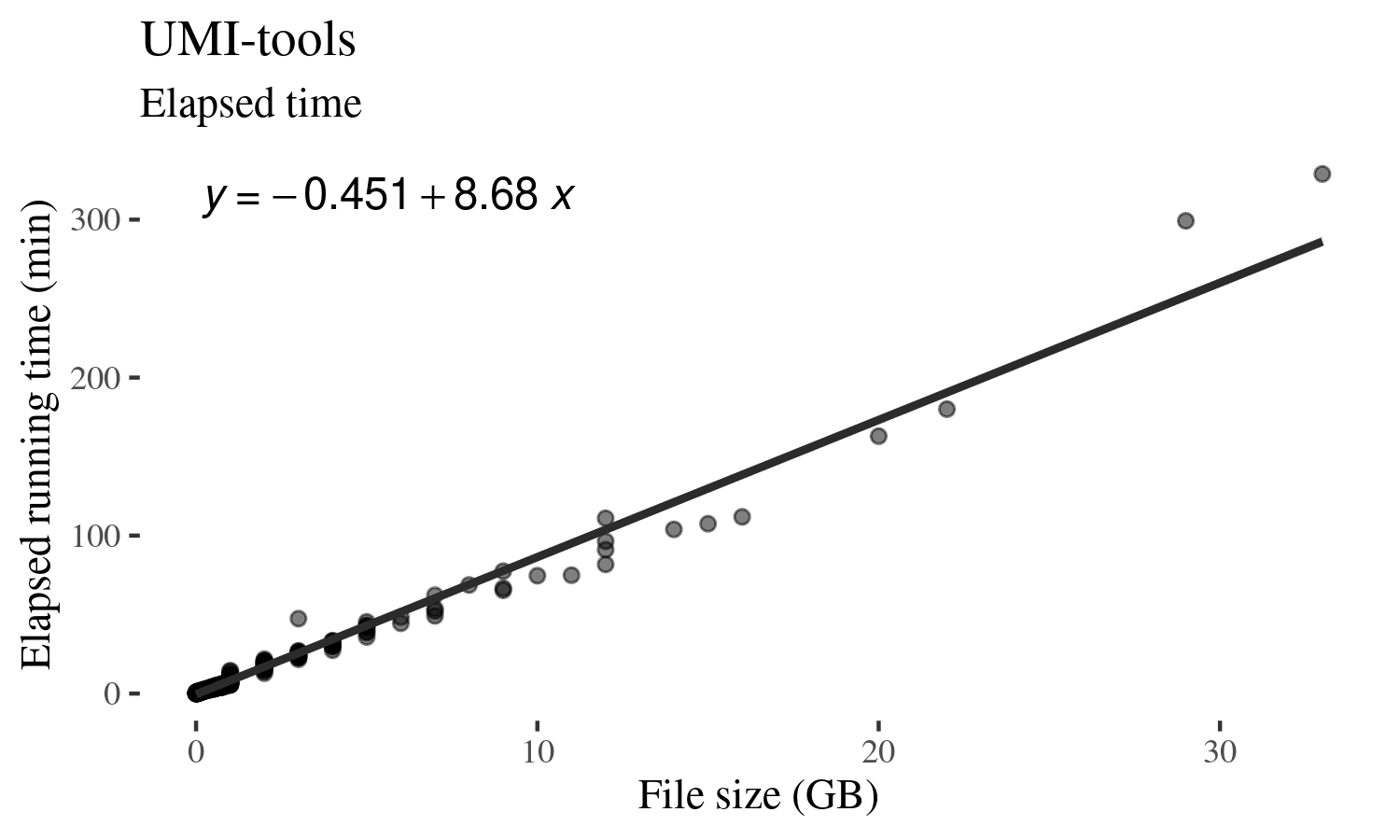
Similarly, the time it takes to de-duplicate a BAM file scales linearly with a much less variation than RAM usage.
Given the information, I am currently requesting a minimum memory of (1GB + 75% of input file size), which seems to serve me well so far.
As for computation time to request, I request 20 minutes per GB of input file plus a baseline of 30 minutes (e.g., for 2GB input, I request 70 minutes). This has usually been an overkill so it could be tune down further.
All jobs were run with one core as
umi-tools does not natively support
multithreading yet.
It should be possible to de-duplicate each
chromosome manually if running the whole BAM proves to be intractable though.